
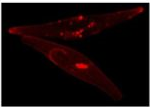
Zeiss Axiovert Fluorescent Microscope
The epifluorescence inverted microscope is used to visualize phytoplankton and other microbes. It is equipped with several imaging modalities, including plastics-compatible DIC, brightfield, and fluorescence with filter sets designed for DAPI, FITC/GFP, chlorophyll and rhodamine imaging. In this image, the diatom Phaeodactylum tricornutum expresses a protein modified with a red fluorescent tag. This iron stress induced protein (ISIP2A) is involved in iron uptake via a pathway similar to that used in human red blood cells (Nature). For high resolution images, we collaborate with Prof. Greg Weber and the Rutgers-Newark Imaging Center.
Trace Metal Clean Lab
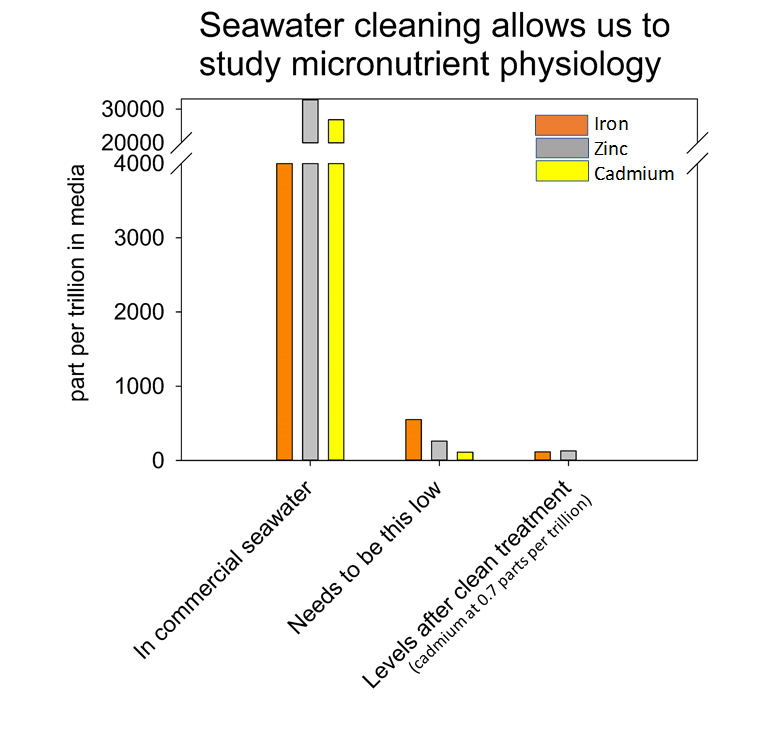
Left panel: Solare recirculating HEPA filtered hood protects precious samples from trace contaminants introduced by workers while protecting workers from harmful fumes from acids or organic solvents. Middle panel: Josephine Arewa adjusting column of chelex resin to strip out trace metal contaminants. Right panel: Commercially available salts contain high levels of iron, zinc and cadmium (left on graph), much higher than the lower amounts we need to achieve for experimentation (middle on graph). Passing artificial salts through chelex brings metal contamination to an acceptable level (right on graph).


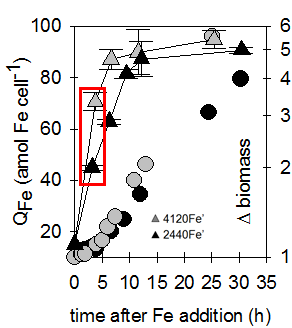
Radioisotope Laboratory
The radioisotope lab allows us to evaluate kinetics of nutrient uptake by phytoplankton and other microbes at low concentrations and high temporal resolution. For example, we can measure exceptionally small but important changes in iron quota in diatoms, as low as 2 attomoles of iron per cell, using 59-Fe radioisotope. After adding high levels of iron back to iron-limited diatoms, we see only about three hours are required to achieve an “iron sufficient” quota of ~ 50 attomoles. Other radionuclides used in our lab include 14-C, 33-P, 54-Mn, 55-Fe, 63-Ni, 65-Zn, as well as tritiated vitamins and 125-I-labeled proteins.
Bio-Rad PDS-1000/HE Biolistics Particle Delivery System
This specialized device is vital for using phytoplankton as a model system for molecular biology applications. It utilizes pressurized helium to launch microscopic gold or tungsten beads coated with DNA at cells with the intention of physically breaking their genome. The cellular DNA repair will on rare occasion incorporate this foreign DNA, leading to a genomic edit.

Graphite Furnace atomic absorption spectrometer
This highly sensitive instrument is used to measure trace metals at the parts per billion or even parts per trillion levels. For example, graduate student David Shire has used this in combination with anion exchange chromatrography and size exclusion HPLC to separate proteins containing the (usually) toxic metal cadmium. This is an optimal approach when small (<30) samples are run at a given time. For higher throughput analysis, we use ICP-MS facilities with collaborator Prof. Robert Sherrell at Marine Sciences

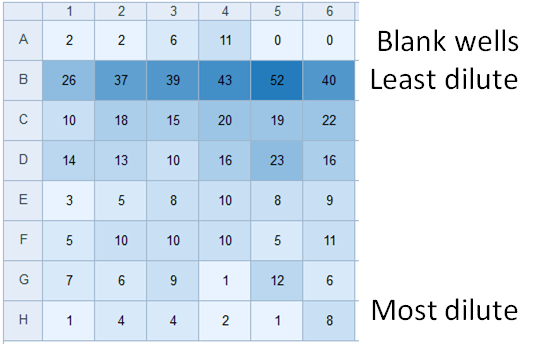
Absorbance/Fluorescence Plate Reader
Unlike fixed-filter plate readers, excitation and emission wavelengths are driven by a monochromator. This permits wavelengths suitable for detection of non-conventional analytes (chlorophyll). The bottom panel show output output from an experiment to quantify survival of diatoms after cryopreservation (numbers indicate relative chlorophyll fluorescence signature). Resurrected cells are serially diluted from row B to row H and survivability is calculated using the most probable number method. Individual wells are scored as containing living cells if chl a fluorescence exceeds background (row A) three weeks after resurrection. Led by undergraduate Josephine Arewa.
Multisizer 3 Coulter Counter
This device simultaneously counts and measures cellular biovolume of cultures, capable of enumerating and sizing up to 1 million cells per milliliter in 30 seconds.

Q Exactive™ HF Hybrid Quadrupole-Orbitrap™



